
pipeML
pipeML.Rmd
library(pipeML)
library(caret)
#> Loading required package: ggplot2
#> Loading required package: lattice
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, unionThis tutorial demonstrates how to use the pipeML package
for training and testing machine learning models. It introduces the main
functions of the pipeline and guides you through additional functions
for visualization.
Load data
raw.counts = pipeML::raw.counts
traitData = pipeML::traitData
deconvolution = pipeML::deconvolutionFeature selection
Apply repeated feature selection using the Boruta algorithm:
deconvolution$target = as.factor(traitData$Best.Confirmed.Overall.Response)
res_boruta <- feature.selection.boruta(
data = deconvolution,
iterations = 5,
fix = FALSE,
doParallel = FALSE,
threshold = 0.8,
file_name = "Test",
return = FALSE
)
#> Running 5 iterations of the Boruta algorithm
#>
#> Keeping only features confirmed in more than 0.8 of the times for training...............................................................
#> If you want to consider also tentative features, please specify tentative = TRUE in the parameters.Inspect the results:
head(res_boruta$Matrix_Importance)
#> NULL
cat("Confirmed features:\n", res_boruta$Confirmed)
#> Confirmed features:
cat("Tentative features:\n", res_boruta$Tentative)
#> Tentative features:To further assess tentative features, set fix = TRUE to
rerun the selection and confirm or reject them:
res_boruta <- feature.selection.boruta(
data = deconvolution,
iterations = 5,
fix = TRUE,
doParallel = FALSE,
threshold = 0.8,
file_name = "Test",
return = FALSE
)For faster execution, enable parallelization (ensure parameters
doParallel and workers are set):
res_boruta <- feature.selection.boruta(
data = deconvolution,
iterations = 10,
fix = FALSE,
doParallel = FALSE,
workers = 2,
threshold = 0.8,
file_name = "Test",
return = FALSE
)Train Machine Learning models
Train and tune models using repeated stratified k-fold cross-validation:
deconvolution = pipeML::deconvolution
traitData = pipeML::traitData
res <- compute_features.training.ML(features_train = deconvolution,
target_var = traitData$Best.Confirmed.Overall.Response,
task_type = "classification",
trait.positive = "CR",
metric = "AUROC",
stack = FALSE,
k_folds = 2,
n_rep = 2,
LODO = FALSE,
file_name = "Test",
ncores = 2,
return = FALSE)Access the best-trained model:
res[[1]]$ModelView all trained and tuned machine learning models:
res[[1]]$ML_Models
knitr::include_graphics("figures/Training.png")
Figure 1. Models training performance.
To apply model stacking, set stack = TRUE:
res <- compute_features.training.ML(features_train = deconvolution,
target_var = traitData$Best.Confirmed.Overall.Response,
task_type = "classification",
trait.positive = "CR",
metric = "AUROC",
stack = TRUE,
k_folds = 2,
n_rep = 2,
LODO = FALSE,
file_name = "Test",
ncores = 2,
return = FALSE)Inspect the base models used in stacking:
res$Model$Base_modelsAccess the meta-learner:
res$Model$Meta_learnerModel prediction
After training, users can predict on new data by using the
compute_prediction() function. You can specify which metric
to maximize when determining the optimal classification threshold.
Supported values for maximize include: “Accuracy”, “Precision”,
“Recall”, “Specificity”, “Sensitivity”, “F1”, and “MCC”.
pred = pipeML::compute_prediction(model = res,
test_data = testing_set,
target_var = traitData$Best.Confirmed.Overall.Response,
trait.positive = "CR",
file.name = "Test",
maximize = "Accuracy",
return = F)If return = TRUE, compute_prediction() will
saved the RO and PR curves in the Results/ directory. It
can also be retrieved with the following function
get_curves(pred$Metrics, spec = "Specificity",
sens = "Sensitivity", reca = "Recall",
prec = "Precision", color = "Cohort",
auc_roc = pred$AUC$AUROC,
auc_prc = pred$AUC$AUPRC,
file.name = "Test")To enable parallelization, specify the number of cores via the
ncores parameter (default is
detectCores() - 1).
res <- compute_features.training.ML(features_train = deconvolution,
target_var = traitData$Best.Confirmed.Overall.Response,
task_type = "classification",
trait.positive = "CR",
metric = "AUROC",
stack = TRUE,
k_folds = 3,
n_rep = 3,
LODO = FALSE,
file_name = "Test",
ncores = 2,
return = FALSE)Train and predict using the machine learning models
If you already have a separate testing dataset, you can directly train the model and perform predictions using the following function. You can also choose to optimize a specific metric to select a decision threshold and return detailed prediction performance metrics such as Accuracy, Sensitivity, Specificity, F1 score, MCC, Recall, and Precision.
deconvolution = pipeML::deconvolution
data_train = deconvolution[1:100,]
data_test = deconvolution[!rownames(deconvolution) %in% rownames(data_train),]
res <- compute_features.ML(features_train = data_train,
features_test = data_test,
clinical = traitData,
task_type = "classification",
trait = "Best.Confirmed.Overall.Response",
trait.positive = "CR",
metric = "AUROC",
stack = FALSE,
k_folds = 3,
n_rep = 3,
LODO = FALSE,
file_name = "Test",
maximize = "F1",
return = FALSE)Access the prediction metrics:
head(res$Prediction_metrics)To compute variable importance:
importance = compute_variable.importance(res$Model,
stacking = FALSE,
n_cores = 2)And to visualize SHAP values:
plot_shap_values(importance, "glm", "shap_plot")
knitr::include_graphics("figures/var_importance.png")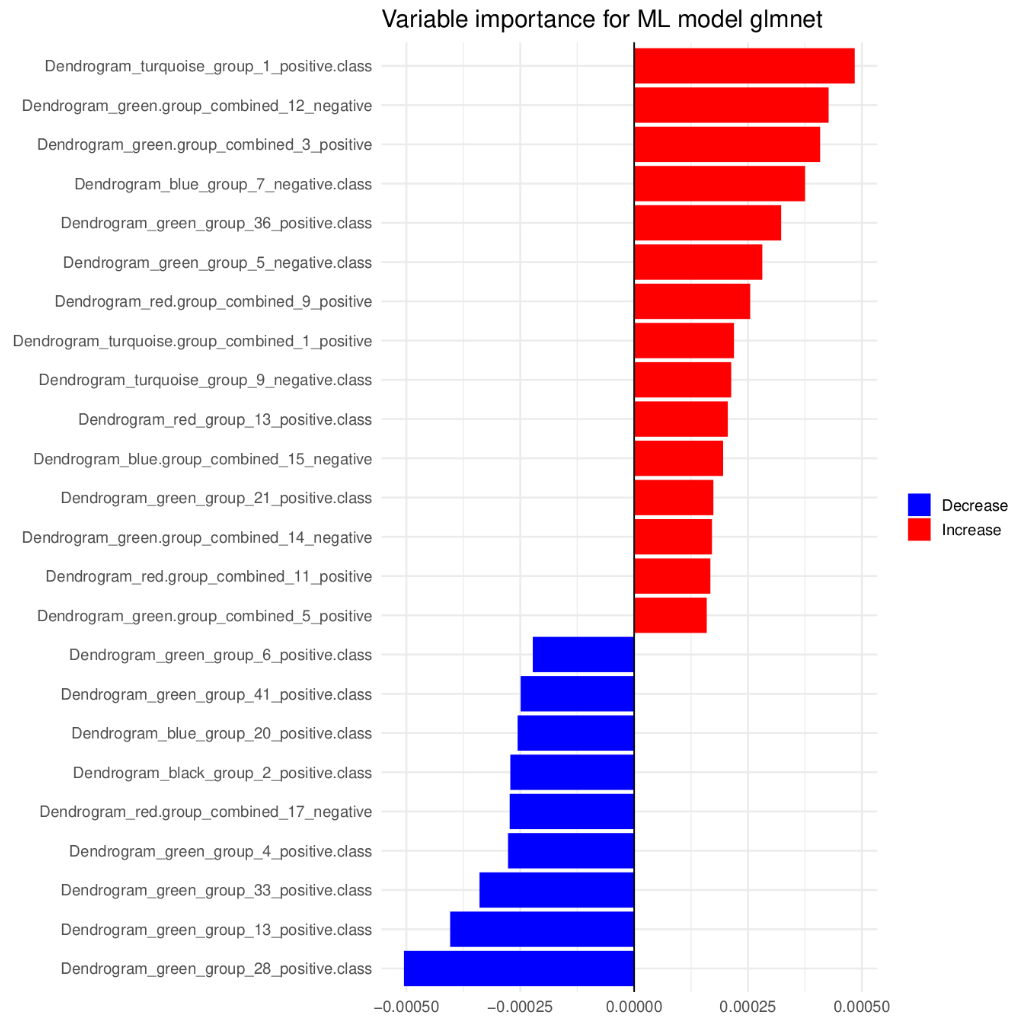
Figure 2. Variable importance.
Leaving one dataset out (LODO) analysis
If your data includes features from multiple cohorts, pipeML provides
a flexible approach to perform Leave-One-Dataset-Out (LODO) analysis.
This is achieved by applying k-fold stratified sampling across batches,
ensuring that each fold preserves the batch structure while maintaining
class balance. For this set LODO = TRUE and provided column
name containing the batch information in the batch_var
variable
Below, we demonstrate how to perform a LODO analysis using simulated datasets:
Simulate datasets from different batches
set.seed(123)
# Simulate traitData with 3 cohorts
traitData <- data.frame(
Sample = paste0("Sample", 1:90),
Response = sample(c("R", "NR"), 90, replace = TRUE),
Cohort = rep(paste0("Cohort", 1:3), each = 30),
stringsAsFactors = FALSE
)
rownames(traitData) <- traitData$Sample
# Simulate counts matrix
counts_all <- matrix(rnorm(1000 * 90), nrow = 1000, ncol = 90)
rownames(counts_all) <- paste0("Gene", 1:1000)
colnames(counts_all) <- traitData$Sample
# Simulate some example features
features_all <- matrix(runif(90 * 15), nrow = 90, ncol = 15)
rownames(features_all) <- traitData$Sample
colnames(features_all) <- paste0("Feature", 1:15)Perform LODO analysis
prediction = list()
i = 1
for (cohort in unique(traitData$Cohort)) {
# Test cohort
traitData_test = traitData %>%
filter(Cohort == cohort)
counts_test = counts_all[,colnames(counts_all)%in%rownames(traitData_test)]
features_test = features_all[rownames(features_all)%in%rownames(traitData_test),]
# Train cohort
traitData_train = traitData %>%
filter(Cohort != cohort)
counts_train = counts_all[,colnames(counts_all)%in%rownames(traitData_train)]
features_train = features_all[rownames(features_all)%in%rownames(traitData_train),]
#### ML Training
res = compute_features.training.ML(features_train = features_train,
target_var = traitData_train$Response,
task_type = "classification",
trait.positive = "R",
metric = "AUROC",
stack = T,
k_folds = 2,
n_rep = 3,
LODO = TRUE,
batch_var = traitData_train$Cohort,
ncores = 2,
return = F)
## Feature importance
importance = compute_variable.importance(res, stacking = T, n_cores = 2)
#### ML predicting
#### Testing
pred = compute_prediction(model = res,
test_data = features_test,
target_var = traitData_test$Response,
trait.positive = "R",
stack = TRUE,
return = F)
pred[["Importance"]] = importance
#### Save results
prediction[[i]] = pred
names(prediction)[i] = cohort
i = i + 1
}For plotting we will make use of our get_curves function
adapted in a case for multiple cohorts.
metrics = list()
auroc = c()
auprc = c()
for (i in 1:length(prediction)) {
metrics[[i]] = prediction[[i]]$Metrics %>%
dplyr::mutate(Cohort = names(prediction)[i],
AUROC = rep(prediction[[i]]$AUC$AUROC, nrow(prediction[[i]]$Metrics)),
AUPRC = rep(prediction[[i]]$AUC$AUPRC, nrow(prediction[[i]]$Metrics))) %>%
tibble::rownames_to_column("Samples")
}
metrics = do.call(rbind, metrics)
metrics$Cohort = as.factor(metrics$Cohort)
get_curves(metrics, "Specificity", "Sensitivity",
"Recall", "Precision", "Cohort",
metrics$AUROC, metrics$AUPRC, "Test")
knitr::include_graphics("figures/ROC.png")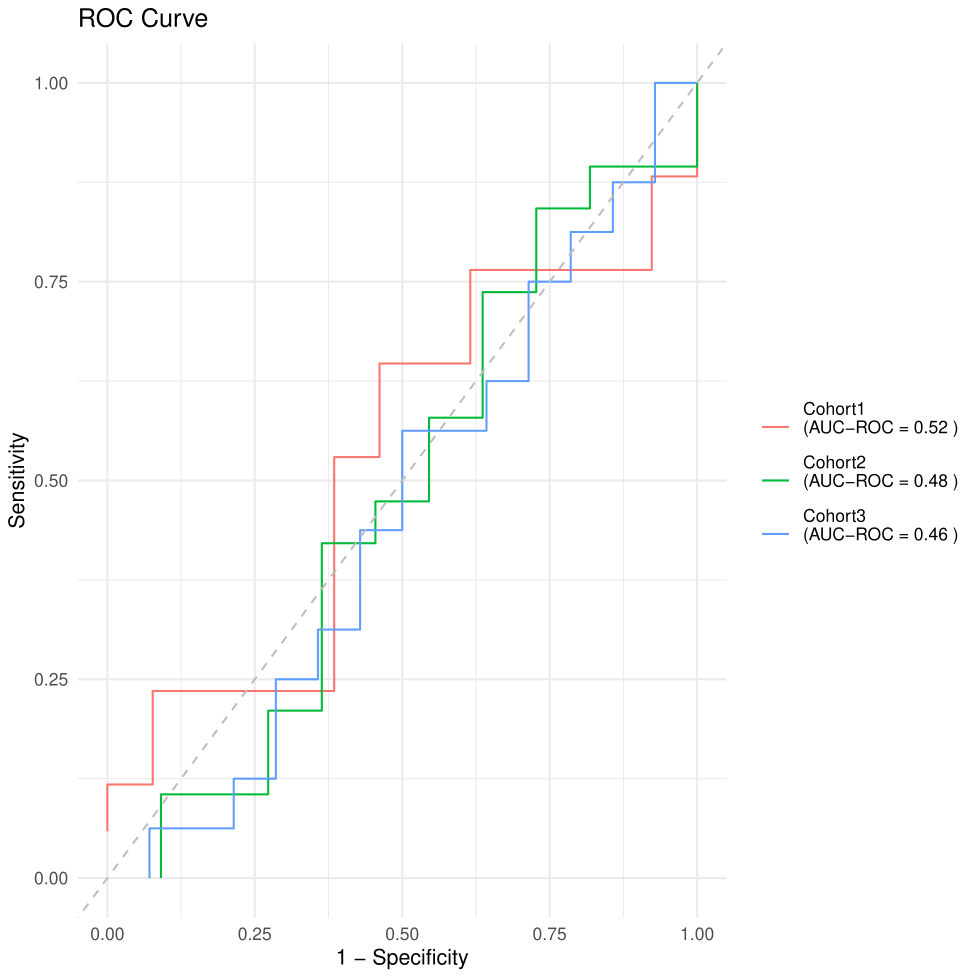
Figure 3. RO curves.
knitr::include_graphics("figures/PR.png")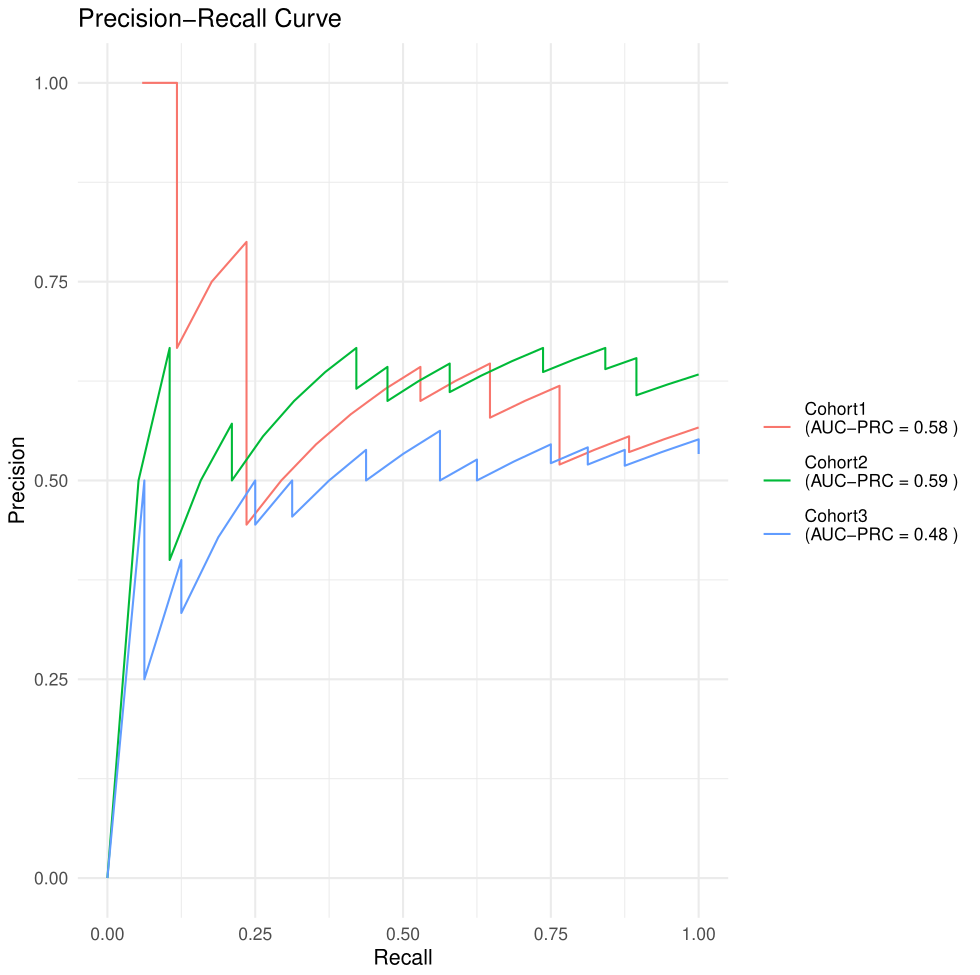
Figure 4. PR curves.
Custom k-fold cross validation
In certain use cases it is essential to carefully design how the
training and test splits are created to prevent data leakage.
pipeML supports the use of custom fold construction
functions via the fold_construction_fun argument in the
compute_features.training.ML() function. This allows you to
preprocess your features within each fold, ensuring that information
from the test set is never used during training.
Why use custom folds?
By default, cross-validation splits are performed directly on the feature matrix. However, if your features are generated through a process that uses correlations (e.g., clustering, dimensionality reduction, aggregation), gene set enrichment or other features that are sample-dependent, applying that process to the full dataset before splitting can leak test-set information into the training set, overfitting the models.
Using a custom fold construction function ensures that: - Test samples are projected onto the learned space without influencing it. - Fold-level processing mimics how the model would behave on truly unseen data.
Below you can find an example of a custom fold function created to compute TF activity in samples using the viper algorithm. This approach is based on a TF-target set enrichment analysis similar to GSEA and thus, the scores will be sample-dependent. It needs normalized counts as input.
# These parameters are taking care inside pipeML automatically once `fold_construction_fun` is set. Be sure to not change the parameteres names.
#' @param data Data from which the custom function will be performed
#' @param folds A list of integer vectors indicating row indices for the training set in each fold.
#'
prepare_TFs_folds <- function(data, folds) {
processed_folds <- list()
for (i in seq_along(folds)) {
train_idx <- folds[[i]]
test_idx <- setdiff(seq_len(nrow(data)), train_idx)
## Subset data
train_data <- data[train_idx, , drop = FALSE]
obs_train <- train_data$target
train_data$target <- NULL
## Run TFs once
train_processed <- compute_TF_activity(RNA_tpm = t(train_data)) ##################################################### User can change this part
colnames(train_processed) <- gsub("[- +]", ".", colnames(train_processed)) ## Just to change special characters (ignore in other cases)
train_cell_data <- train_processed %>%
dplyr::mutate(target = obs_train)
## Prepare test data using trained info
obs_test <- data$target[test_idx]
test_data <- data[test_idx, , drop = FALSE]
test_data$target <- NULL
test_data <- compute_TF_activity(RNA_tpm = t(test_data)) ##################################################### User can change this part
colnames(test_data) <- gsub("[- +]", ".", colnames(test_data)) ## Just to change special characters (ignore in other cases)
processed_folds[[i]] <- list(
train_data = train_cell_data,
test_data = test_data,
obs_test = obs_test,
rowIndex = test_idx,
fold_name = names(folds)[i]
)
}
# Run TFs on the full training set
obs_train = data$target
data$target = NULL
train_processed_final <- compute_TF_activity(RNA_tpm = t(data)) ##################################################### User can change this part
colnames(train_processed_final) <- gsub("[- +]", ".", colnames(train_processed_final)) ## Just to change special characters (ignore in other cases)
# Get features
train_cell_data_final <- train_processed_final %>%
dplyr::mutate(target = obs_train)
custom_output = train_processed_final
return(list(processed_folds, train_cell_data_final, custom_output))
}In order to create your own custom fold function, user can follow the
same structure defined above and change the part highlighted in the
code. Once the function is ready you can provide it into the main
compute_features.training.ML() function via the
fold_construction_fun argument.
## counts_train: Normalized counts to compute the features from the training samples
## traitData_train: Target variable from the training samples
res = pipeML::compute_features.training.ML(as.data.frame(t(counts_train)),
traitData_train$Response,
trait.positive = "R",
metric = "AUROC",
stack = F, k_folds = 5,
n_rep = 10, LODO = F,
ncores = 2, return = F,
fold_construction_fun = prepare_TFs_folds)In order to predict, user needs to calculate the same type of features on the test set
## counts_test: Normalized counts to compute the features from the testing samples
tf_activities_test <- compute_TF_activity(RNA_tpm = counts_test)
# Predict in TFs
pred = compute_prediction(res, tf_activities_test, traitData_test$Response, trait.positive = "R",
stack = FALSE, maximize = "Accuracy", return = F)What if fold_construction_fun has multiple
parameters?
Sometimes the feature construction function requires additional
arguments. For example, compute_TF_activity() may need the
TF collection, a minimum target size, and whether TFs should be pruned.
These can be exposed in your custom function:
tfs = compute.TFs.activity(counts.norm, TF.collection = "CollecTRI", min_targets_size = 5, tfs.pruned = FALSE)Then the custom function needs to specify these parameters and their values. It will look like this:
prepare_TFs_folds <- function(data, folds, TF.collection = "CollecTRI", min_targets_size = 5, tfs.pruned = FALSE) {
processed_folds <- list()
for (i in seq_along(folds)) {
train_idx <- folds[[i]]
test_idx <- setdiff(seq_len(nrow(data)), train_idx)
## Subset data
train_data <- data[train_idx, , drop = FALSE]
obs_train <- train_data$target
train_data$target <- NULL
##################################################### User can change this part
train_processed <- compute_TF_activity(RNA_tpm = t(train_data),
TF.collection = "CollecTRI",
min_targets_size = 5,
tfs.pruned = FALSE)
colnames(train_processed) <- gsub("[- +]", ".", colnames(train_processed)) ## Just to change special characters (ignore in other cases)
train_cell_data <- train_processed %>%
dplyr::mutate(target = obs_train)
## Prepare test data using trained info
obs_test <- data$target[test_idx]
test_data <- data[test_idx, , drop = FALSE]
test_data$target <- NULL
##################################################### User can change this part
test_data <- compute_TF_activity(RNA_tpm = t(test_data),
TF.collection = "CollecTRI",
min_targets_size = 5,
tfs.pruned = FALSE)
colnames(test_data) <- gsub("[- +]", ".", colnames(test_data)) ## Just to change special characters (ignore in other cases)
processed_folds[[i]] <- list(
train_data = train_cell_data,
test_data = test_data,
obs_test = obs_test,
rowIndex = test_idx,
fold_name = names(folds)[i]
)
}
# Run TFs on the full training set
obs_train = data$target
data$target = NULL
##################################################### User can change this part
train_processed_final <- compute_TF_activity(RNA_tpm = t(data),
TF.collection = "CollecTRI",
min_targets_size = 5,
tfs.pruned = FALSE)
colnames(train_processed_final) <- gsub("[- +]", ".", colnames(train_processed_final)) ## Just to change special characters (ignore in other cases)
# Get features
train_cell_data_final <- train_processed_final %>%
dplyr::mutate(target = obs_train)
custom_output = train_processed_final
return(list(processed_folds, train_cell_data_final, custom_output))
}When calling compute_features.training.ML(), these fixed
arguments are passed via fold_construction_args_fixed:
res = pipeML::compute_features.training.ML(as.data.frame(t(counts_train)),
traitData_train$Response,
task_type = "classification",
trait.positive = "R",
metric = "AUROC",
stack = F, k_folds = 5,
n_rep = 10, LODO = F,
ncores = 2, return = F,
fold_construction_fun = prepare_TFs_folds,
fold_construction_args_fixed = list(TF.collection = "CollecTRI",
min_targets_size = 5,
tfs.pruned = FALSE))Tunable hyperparameters during custom function
There will be cases when user will have parameters that can change
value and thus, affect the performance during training. pipeML
implements a custom hyperparameter tuning for these type of cases where
the goal is to find which value of a certain parameter maximizes the
performance of the model. Briefly, it takes a grid of values propose by
the user and compute custom k-folds for each combination of values,
testing the performance across different ML models and their
corresponding hyperparameters. It then returns the optimized parameters
that return the best performance across a certain number of iterations
defined by the user. For this, user needs to defined the parameter
fold_construction_args_tunable with the different values
that should be test. Here we want to test the performance using 5, 10,
15, 20 as different values for this parameter.
res = pipeML::compute_features.training.ML(as.data.frame(t(counts_train)),
traitData_train$Response,
task_type = "classification",
trait.positive = "R",
metric = "AUROC",
stack = F, k_folds = 5,
n_rep = 10, LODO = F,
ncores = 2, return = F,
fold_construction_fun = prepare_TFs_folds,
fold_construction_args_fixed = list(TF.collection = "CollecTRI", tfs.pruned = FALSE),
fold_construction_args_tunable = list(min_targets_size = c(5, 10, 15, 20)))For this to work, your custom function must accept a
bestune argument, which is used internally to inject the
optimized parameter values found during the tuning step.
When bestune is NULL, the function assumes that tuning
has not yet been performed. A grid of candidate parameter values
(defined in fold_construction_args_tunable) is generated.
For each fold, the function iterates through all combinations of
parameter values and recomputes the features. This exploration step is
parallelized across folds using foreach and doParallel, allowing
multiple folds to be processed simultaneously. Parallelization reduces
runtime considerably when the parameter grid or number of folds is
large.
When bestune is not NULL, it means the tuning process
has already been completed. The optimized parameter values are extracted
from the bestune object. Features are then recomputed once on the full
training dataset using these tuned parameters. This ensures the final
model is trained with the best parameter setting identified during
cross-validation.
In summary, the bestune argument acts as a control
switch:
- NULL → run parameter search with parallelized fold-level evaluation.
- non-NULL → lock in the tuned parameter values and rebuild the features for final training.
This design allows a single custom fold-construction function to handle both hyperparameter tuning (exploration, parallelized) and final model preparation (exploitation, single optimized run).
prepare_TFs_folds <- function(data, folds, TF.collection = "CollecTRI", min_targets_size = 5, tfs.pruned = FALSE, bestune = NULL){
if(!is.null(bestune)){
obs_train = data$target
data$target = NULL
best_params <- bestune %>%
dplyr::select(min_targets_size) ### Here you should put the hyperparameters that you want to tune
train_processed_final <- compute_TF_activity(RNA_tpm = t(test_data),
TF.collection = "CollecTRI",
min_targets_size = best_params$min_targets_size, ### Here you replace by the value of the optimized parameter
tfs.pruned = FALSE)
colnames(train_processed_final) <- gsub("[- +]", ".", colnames(train_processed_final)) ## Just to change special characters (ignore in other cases)
# Get cell group features
train_cell_data_final <- train_processed_final %>%
dplyr::mutate(target = obs_train)
custom_output = train_processed_final
res = list(train_cell_data_final, custom_output, best_params)
}else{ ### Means best tune need to be find
#### Here put all the parameters you want to tune
custom_grid <- expand.grid(
min_targets_size = min_targets_size,
)
### Implement parallelization
if(is.null(ncores) == TRUE){
ncores = parallel::detectCores() - 2
}
cl <- parallel::makeCluster(ncores)
doParallel::registerDoParallel(cl)
# Parallelize over folds
processed_folds <- foreach::foreach(i = seq_along(folds), .packages = c("dplyr")) %dopar% {
cat("Starting fold", names(folds)[i], "\n")
train_idx <- folds[[i]]
test_idx <- setdiff(seq_len(nrow(data)), train_idx)
## Subset data
train_data <- data[train_idx, , drop = FALSE]
train_coldata <- coldata[train_idx, , drop = FALSE]
obs_train <- train_data$target
train_data$target <- NULL
fold_results <- vector("list", nrow(custom_grid))
for (j in seq_len(nrow(custom_grid))) {
##################################################### User can change this part
train_processed <- compute_TF_activity(RNA_tpm = t(train_data),
TF.collection = "CollecTRI",
min_targets_size = custom_grid$min_targets_size[j], ### Here will iterate across params values
tfs.pruned = FALSE)
colnames(train_processed) <- gsub("[- +]", ".", colnames(train_processed)) ## Just to change special characters (ignore in other cases)
train_cell_data <- train_processed %>%
dplyr::mutate(target = obs_train)
## Prepare test data using trained info
obs_test <- data$target[test_idx]
test_data <- data[test_idx, , drop = FALSE]
test_data$target <- NULL
##################################################### User can change this part
test_data <- compute_TF_activity(RNA_tpm = t(test_data),
TF.collection = "CollecTRI",
min_targets_size = custom_grid$min_targets_size[j], ### Here will iterate across params values
tfs.pruned = FALSE)
colnames(test_data) <- gsub("[- +]", ".", colnames(test_data)) ## Just to change special characters (ignore in other cases)
fold_results[[j]] <- list(
train_data = train_cell_data,
test_data = test_data,
obs_test = obs_test,
rowIndex = test_idx,
fold_name = names(folds)[i],
params = custom_grid[j, ]
)
}
fold_results
}
parallel::stopCluster(cl) # stop the cluster after parallel execution
unregister_dopar() #Stop Dopar from running in the background
res <- processed_folds
}
return(res)
}