This tutorial demonstrates how to use the multideconv
package and explains the main functions of the pipeline for deconvolving
RNA-seq data.
Deconvolution with default methods
The basic function is to perform cell type deconvolution using six
default methods (quanTIseq, DeconRNASeq,
CIBERSORTx, EpiDISH, DWLS,
MOMF) and nine default signatures (see paper Hurtado
et al., 2025). The function accepts either raw counts or
TPM-normalized counts as input (with genes as SYMBOL).
NOTE: If you plan to use CIBERSORTx,
you must provide your credentials (see README for details). The
resulting deconvolution matrix is automatically saved in the
Results/ directory.
The output includes all combinations of deconvolution features, method-signature-cell type.
bulk = multideconv::raw_counts
deconv = compute.deconvolution(raw.counts = bulk,
methods = c("Quantiseq", "Epidish",
"DeconRNASeq", "DWLS","MOMF"),
normalized = TRUE,
return = TRUE,
file_name = "Tutorial")To exclude specific methods or signatures, use the methods or signatures_exclude arguments:
deconv = compute.deconvolution(raw.counts = bulk,
methods = c("Quantiseq", "DeconRNASeq"),
normalized = TRUE,
signatures_exclude = "BPRNACan",
return = TRUE,
file_name = "Tutorial")To speed up computation, multideconv supports
parallelization. Set doParallel = TRUE and specify the
number of workers based on your system’s resources:
deconv = compute.deconvolution(raw.counts = bulk,
methods = "DWLS",
normalized = TRUE,
return = TRUE,
file_name = "Tutorial",
doParallel = TRUE,
workers = 3)Cell type signatures
In order to access the default signatures multideconv
provides, you can do the following:
To list all signatures
path <- system.file("signatures/", package = "multideconv")
list.files(path)
#> [1] "BPRNACan.txt" "BPRNACan3DProMet.txt"
#> [3] "BPRNACanProMet.txt" "CBSX-HNSCC-scRNAseq.txt"
#> [5] "CBSX-Melanoma-scRNAseq.txt" "CBSX-NSCLC-PBMCs-scRNAseq.txt"
#> [7] "CCLE-TIL10.txt" "LM22.txt"
#> [9] "MCPcounter" "TIL10.txt"To access a specific signature
signature = read.delim(paste0(path, "CBSX-Melanoma-scRNAseq.txt"))
head(signature)
#> NAME Malignant Endothelial.cells CAF T.cells.CD8 NK.cells Macrophages
#> 1 A2M 272.91408 212.14093 1.00000 1.00000 1.0000 700.2033
#> 2 A4GALT 1.00000 67.08075 1.00000 1.00000 1.0000 1.0000
#> 3 ABCA1 1.00000 84.22255 1.00000 1.00000 1.0000 128.7594
#> 4 ABCB1 1.00000 1.00000 1.00000 1.00000 133.5748 1.0000
#> 5 ABCB5 117.86677 1.00000 1.00000 19.94351 1.0000 1.0000
#> 6 ABCB6 49.62804 1.00000 41.38285 1.00000 1.0000 1.0000
#> T.cells.CD4 B.cells
#> 1 1 1
#> 2 1 1
#> 3 1 1
#> 4 1 1
#> 5 1 1
#> 6 1 1Single-cell metacell construction
If single-cell data is available, we recommend generating metacells to reduce computation time and prevent session crashes. Deconvolution methods that rely on single-cell data can be computationally intensive, especially with large matrices. We suggest using a maximum of 20k cells; if your object exceeds this size, creating metacells is strongly advised. However, if your computational resources are sufficient to handle the full single-cell dataset, you may skip this step.
We adapted functions from the R package hdWGCNA (Morabito et al. (2023); Langfelder and Horvath (2008)) for the construction of metacells using the KNN algorithm.
sc_object: Normalized gene expression matrix with genes as rows and cells as columns
labels_column: Vector of cell annotations
samples_column: Vector of sample IDs for each cell
exclude_cells: Vector specifying which cell types to ignore during metacell construction (default is NULL)
min_cells: Minimum number of cells required to construct metacells in a group
k: Number of nearest neighbors used for the KNN algorithm
max_shared: Maximum number of cells shared between two metacells
n_workers: Number of cores to use for parallelizing metacell construction
min_meta: Minimum number of metacells required for a cell type to be retained
Because of space limitations, we have not included a complete
single-cell object in this tutorial. However, users are expected to
provide their own single-cell data and supply it to the
sc_object parameter in the function call.
metacells = create_metacells(sc_object,
labels_column = cell_labels,
samples_column = sample_labels,
exclude_cells = NULL,
min_cells = 50,
k = 15,
max_shared = 15,
n_workers = 4,
min_meta = 10)Second-generation deconvolution methods
Once the single-cell data is prepared, users can supplement the
default deconvolution methods with second-generation approaches such as
AutogeneS, BayesPrism, Bisque,
CPM, MuSic, and SCDC. These
methods learn cell-type signatures directly from annotated single-cell
RNA-seq data, rather than relying on predefined static signatures (Dietrich et al. (2024)), to deconvolve
bulk RNA-seq profiles.
sc_deconv: Boolean indicating whether to run second-generation methods
sc_matrix: Normalized single-cell gene expression matrix
sc_metadata: Dataframe containing single-cell metadata
cell_annotations: Vector of cell type labels
cell_samples: Vector of sample IDs
name_sc_signature: Name to assign to the resulting signature
metacell_obj = multideconv::metacells_data
metacell_metadata = multideconv::metacells_metadata
head(metacell_obj[1:5,1:5])
#> B cells#Sample_15002_393 CD4+ T cells#Sample_14958_193
#> AL627309.1 0 0
#> AL732372.1 0 0
#> AC114498.1 0 0
#> FAM87B 0 0
#> LINC00115 0 0
#> Plasma B cells#Sample_15467_117 CD4+ T cells#Sample_12889_54
#> AL627309.1 0.00000000 0.00000000
#> AL732372.1 0.00000000 0.00000000
#> AC114498.1 0.00000000 0.00000000
#> FAM87B 0.00000000 0.00000000
#> LINC00115 0.01454298 0.05538857
#> B cells#Sample_11817_97
#> AL627309.1 0
#> AL732372.1 0
#> AC114498.1 0
#> FAM87B 0
#> LINC00115 0
head(metacell_metadata)
#> orig.ident nCount_RNA nFeature_RNA
#> B cells#Sample_15002_393 B cells#Sample 2085.014 7927
#> CD4+ T cells#Sample_14958_193 CD4+ T cells#Sample 1699.248 6390
#> Plasma B cells#Sample_15467_117 Plasma B cells#Sample 1397.391 9314
#> CD4+ T cells#Sample_12889_54 CD4+ T cells#Sample 1798.458 7164
#> B cells#Sample_11817_97 B cells#Sample 1944.183 7765
#> CD8+ T cells#Sample_11522_665 CD8+ T cells#Sample 2317.216 7862
#> cells_merged
#> B cells#Sample_15002_393 CTGTTTATCGCCATAA-1,TGCTGCTTCGGAGCAA-1,ACTTTCATCGACAGCC-1,GCTGCAGTCATTGCGA-1,CGTAGGCGTGGCTCCA-1,TCAGCTCAGACTAAGT-1,CACCAGGCACCAGCAC-1,CATCAAGTCAGCGATT-1,CGATGGCTCGTTACGA-1,CAGTCCTGTGTGAATA-1,CTTGGCTCAAGTCATC-1,CCAATCCAGCCACGTC-1,TTTGGTTCAGATGGCA-1,TGGTTAGAGAGTAATC-1,TAGTTGGGTTATGTGC-1,GACACGCGTCGAGTTT-1,AGGGATGGTACACCGC-1,ACTTACTTCGAGAACG-1,GGCAATTCATTAGGCT-1,GAAACTCAGACTGGGT-1,ATTGGTGGTAGCTTGT-1,TTGAACGAGACGCAAC-1,TGTTCCGGTACTTAGC-1,CAGCTGGCAGGTGGAT-1,GGACATTAGCCCAATT-1,TAAGCGTCAGCCTGTG-1,CTCTAATAGATCTGAA-1,CACACCTAGCCTTGAT-1,CTTACCGAGCGCTCCA-1,CACTCCAGTCGACTGC-1
#> CD4+ T cells#Sample_14958_193 AAGGAGCTCCAAATGC-1,ACACCCTTCTGCGTAA-1,CGGACGTGTAGGCTGA-1,CGACCTTCAAGTCATC-1,TTAGGCACAGGAACGT-1,GTATTCTAGTGTGAAT-1,TCAATCTCAAGAAAGG-1,GGCAATTTCTTGTTTG-1,GCGCAACGTCTAGCCG-1,CCTTACGCATCGACGC-1,TCCCGATAGACAATAC-1,ACAGCTAGTTCCACTC-1,TGAGAGGAGATCTGAA-1,GCGCCAAAGCCACCTG-1,TAAGTGCGTCTCACCT-1,ATGCGATTCGTTACGA-1,CAAGTTGAGCTACCGC-1,TACCTTACATTCTTAC-1,GCATGTATCAGTCAGT-1,GATGCTACATATGGTC-1,CCATGTCTCTGGGCCA-1,TACACGAGTCATTAGC-1,GATTCAGAGCACACAG-1,TTGCCGTAGCAATATG-1,TCGTACCAGCCAACAG-1,CAGTCCTTCGCCAGCA-1,CTACATTCAGTCGATT-1,AGGGTGATCTTGACGA-1,GTCTCGTGTCTCTTAT-1,TCGGGACAGCACACAG-1-1
#> Plasma B cells#Sample_15467_117 GCGCAACAGTACATGA-1,GTCATTTCACGCGAAA-1,ACGATACTCATAAAGG-1,CCCATACCACCAACCG-1,AGCCTAAAGTGTTAGA-1,TGTATTCCAGGGTACA-1,CATATTCTCAACGGCC-1,CACACCTGTCCAACTA-1,CCGTACTTCCGAGCCA-1,GGGACCTTCCAGAAGG-1,ACACCCTGTGTAACGG-1,AGCCTAACAGAAGCAC-1,GCTTCCATCCATGCTC-1,AGTGTCAGTCCTGCTT-1,CGAGCACAGAAACGCC-1,GAACGGATCGGGAGTA-1,TGCCAAACACTCAGGC-1,TACAGTGCAGTGGGAT-1,TATGCCCCAGTATCTG-1,GTCGGGTGTAAGAGGA-1,AAGGCAGCATCTATGG-1,TCGAGGCGTAGCTCCG-1,CTGCTGTTCGCGCCAA-1,CGGACACAGACTACAA-1,CGTGTAAAGCCCAATT-1,CTTTGCGAGGAGTCTG-1,TGCCCTAAGCCAACAG-1,CTCAGAACACCTCGGA-1,CTGCCTAGTGTCTGAT-1,ACACCAATCGACGGAA-1
#> CD4+ T cells#Sample_12889_54 GCGAGAATCGCACTCT-1,GTTACAGTCTTCGAGA-1,CAGTAACGTAACGACG-1,TCGCGTTCAATCCGAT-1,CACCTTGGTGTCGCTG-1,ATTACTCCAAACAACA-1,TACAGTGTCAGTGCAT-1,ACGCAGCGTCTGCCAG-1,TTGTAGGAGCCACGTC-1,CTTGGCTGTTCGTGAT-1,CCACTACAGACACTAA-1,CCCAGTTTCAGGCCCA-1,GAACGGAAGGCACATG-1,CGTAGCGAGAGGGCTT-1,ATTCTACTCGGTTAAC-1,CATGACAGTGGTCCGT-1,TCAGCAAGTGCTTCTC-1,AGCTTGAGTCCTCCAT-1,CATTCGCCATCGATTG-1,TCAGGATAGCACAGGT-1,CCCTCCTTCTGCGTAA-1,AAATGCCTCCTTGACC-1,TACTCGCTCCTATTCA-1,GAAATGATCGCATGGC-1,ACTGCTCTCGGTCTAA-1,AACGTTGTCATAGCAC-1,GTGTGCGGTCTAGGTT-1,GCACTCTCAAGCCGTC-1,CCGTTCACATCCTTGC-1,CCACTACGTCGAGTTT-1
#> B cells#Sample_11817_97 AAACCTGGTACCGGCT-1,ACAGCTAGTAAGTAGT-1,ACGATACAGTACGACG-1,TAAACCGAGGAGTACC-1,GGACGTCGTCTGCAAT-1,TAGCCGGAGGGCATGT-1,CGGACACCACATCCAA-1,GGTATTGTCACTTCAT-1,GGAAAGCAGTACGATA-1,AAACGGGTCGTGGGAA-1,TTTCCTCCAGGCGATA-1,GTCATTTAGGAACTGC-1,CAACCAAGTCCGTTAA-1-1,GTCGTAAGTACAGTGG-1,GTTTCTAAGCCCAATT-1,GACCAATCATTCCTCG-1,CAACCAAGTTCTGAAC-1,AGATCTGAGCAGGTCA-1,GTTCGGGCAGTTCCCT-1,GCAAACTCATGAACCT-1,CTCTGGTGTGATGATA-1,TTATGCTCAAACTGTC-1,GACGTTAGTACCGTTA-1,GGGACCTGTTTGACAC-1,ACAGCCGTCTGCGGCA-1,GGCGTGTTCTAAGCCA-1,CGGACACGTGCTTCTC-1-1,GGTGAAGAGCTCAACT-1,GCTTGAACATCCGTGG-1,GGGAGATAGGGCACTA-1
#> CD8+ T cells#Sample_11522_665 ACACCGGAGCCAGTAG-1-1,GAAATGATCCCACTTG-1,GAGTCCGAGTACGCGA-1,CTGAAACAGTCGCCGT-1,GTCGGGTGTGGTGTAG-1,ACGATGTCAGACGCTC-1,ATCATCTGTCTGCAAT-1,TGCTGCTAGACACTAA-1,TACTTGTTCGCAAGCC-1,CGAGCACAGTTAGGTA-1,GCTTGAATCTACTCAT-1,ATCATCTCAAGACGTG-1,GTGTTAGGTCAACATC-1,CGGGTCAGTAAGCACG-1,GAACCTAAGAGTGAGA-1,TCATTACGTGCAACGA-1,GACGTGCGTATGCTTG-1,TCGTACCTCATGCTCC-1,TGCGCAGTCGCCAGCA-1,CACACTCGTAGCAAAT-1,GATGAAAGTATCAGTC-1,TCATTTGAGGCTATCT-1,CTAACTTTCAACTCTT-1,CTTGGCTCAAAGAATC-1,GACTACATCTGACCTC-1,TTTCCTCTCAACGAAA-1,AAAGCAAGTCAGAAGC-1,CATCAAGTCCGAATGT-1,CGATGGCTCGCAAACT-1,CGAGAAGTCAGTGTTG-1
#> annotated_ct sample
#> B cells#Sample_15002_393 B cells Sample_15002
#> CD4+ T cells#Sample_14958_193 CD4+ T cells Sample_14958
#> Plasma B cells#Sample_15467_117 Plasma B cells Sample_15467
#> CD4+ T cells#Sample_12889_54 CD4+ T cells Sample_12889
#> B cells#Sample_11817_97 B cells Sample_11817
#> CD8+ T cells#Sample_11522_665 CD8+ T cells Sample_11522This function computes cell type deconvolution using the six default
methods (quanTIseq, DeconRNASeq,
EpiDISH, DWLS, MOMF) and
CIBERSORTx (if credentials are provided), along with
second-generation deconvolution approaches. The output includes all
combinations of methods and signatures.
deconv = compute.deconvolution(raw.counts = bulk,
normalized = TRUE,
return = TRUE,
methods = c("Quantiseq", "Epidish", "DeconRNASeq"),
file_name = "Tutorial",
sc_deconv = TRUE,
sc_matrix = metacell_obj,
sc_metadata = metacell_metadata,
methods_sc = c("Autogenes", "BayesPrism",
"Bisque", "CPM", "MuSic", "SCDC"),
cell_label = "annotated_ct",
sample_label = "sample",
name_sc_signature = "Test")To run only the second-generation deconvolution methods based on single-cell data, without using any static cell-type signatures, use the following:
deconv_sc = compute_sc_deconvolution_methods(raw_counts = bulk,
normalized = TRUE,
methods_sc = c("Autogenes", "BayesPrism",
"Bisque", "CPM", "MuSic", "SCDC"),
sc_object = metacell_obj,
sc_metadata = metacell_metadata,
cell_annotations = "annotated_ct",
samples_ids = "sample",
name_object = "Test",
n_cores = 2,
return = TRUE,
file_name = "Tutorial")Pseudo-bulk profiles
To create pseudo-bulk profiles from the original single-cell objects, simulating a bulk RNA-seq dataset, you can use the following function:
NOTE: You can input either your original single-cell object or the metacell object. Just be sure to select the same object when examining the real cell proportions (if needed).
metacells_seurat = Seurat::CreateSeuratObject(metacell_obj, meta.data = metacell_metadata)
#> Warning: Data is of class matrix. Coercing to dgCMatrix.
pseudobulk = create_sc_pseudobulk(metacells_seurat, cells_labels = "annotated_ct", sample_labels = "sample", normalized = TRUE, file_name = "Tutorial")
#> Warning: Layer 'data' is empty
#> Warning: Layer 'scale.data' is empty
#> Aggregating assay 'counts' using 'rowMeans2'.
#> Converting input to matrix.Creating cell type signatures
To create cell type signatures, multideconv uses four
methods: CIBERSORTx, DWLS, MOMF,
and BSeq-SC. You must provide single-cell data as input.
Signatures are saved in the Results/custom_signatures
directory, and returned as a list. From now and after
compute.deconvolution() will use these signatures
additionally to the default ones! So if you would like to have the
deconvolution results based on your new files, make sure to run
compute.deconvolution()
To run BSeq-SC, supply the cell_markers
argument, which should contain the differential markers for each cell
type (these can be obtained using FindMarkers() or
FindAllMarkers() from Seurat).
bulk_pseudo = multideconv::pseudobulk
signatures = create_sc_signatures(metacell_obj,
metacell_metadata,
cells_labels = "annotated_ct",
sample_labels = "sample",
bulk_rna = bulk_pseudo,
cell_markers = NULL,
name_signature = "Test",
methods_sig = c("DWLS", "CIBERSORTx", "MOMF", "BSeqsc"))Cell types signatures benchmark
To validate the generated signatures, we provide a benchmarking
function to compare deconvolution outputs against known cell proportions
(e.g., from single-cell or imaging data). The cells_extra
argument should include any non-standard cell types present in your
ground truth. Make sure cell names match those in the deconvolution
matrix (e.g., use B.cells instead of B cells if that is the naming
convention used - see README for more information).
deconv_pseudo = multideconv::deconvolution
cells_groundtruth = multideconv::cells_groundtruth
benchmark = compute.benchmark(deconv_pseudo,
cells_groundtruth,
cells_extra = c("Mural.cells", "Myeloid.cells"),
corr_type = "spearman",
scatter = FALSE,
plot = TRUE,
pval = 0.05,
file_name = "Tutorial",
width = 10,
height = 15)
#> No id variables; using all as measure variables
#> No id variables; using all as measure variables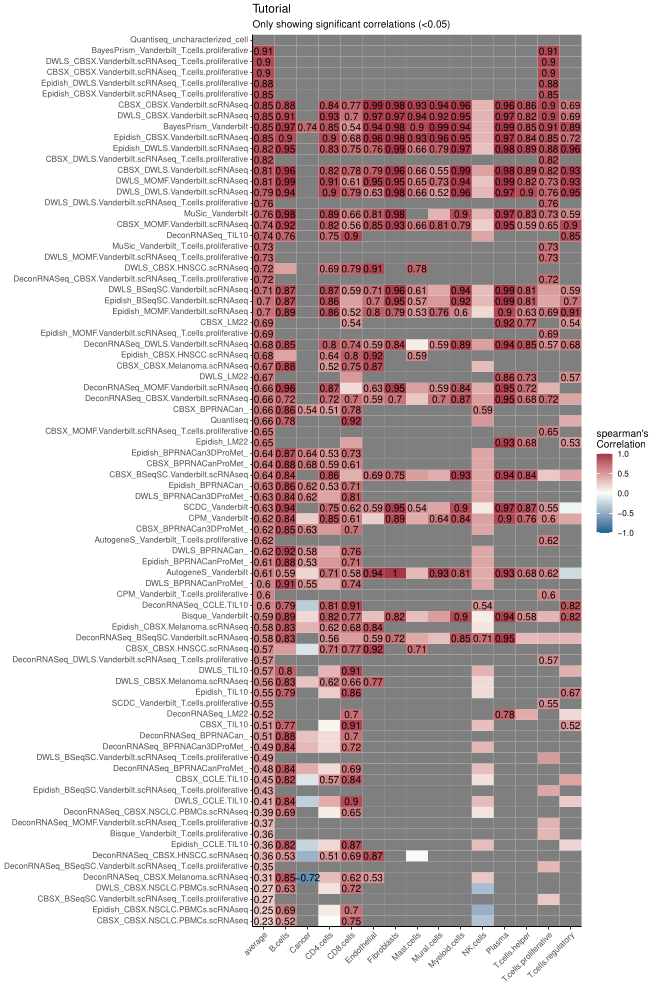
Figure 1. Example of performance of different methods and signature combinations on the pseudo bulk.
Cell type processing
Deconvolution analysis reduces the dimensionality and heterogeneity
of the deconvolution results. It uses the cell type processing algorithm
described in the paper Hurtado
et al., 2025. It returns the cell type subgroups composition and the
reduced deconvolution matrix, saved in the Results/
directory.
deconvolution: Matrix of raw deconvolution results (output of
compute.deconvolution())corr: Minimum correlation threshold to group features
seed: Random seed for reproducibility
return: Whether to return results and save output files to the
Results/directory
deconv_bulk = multideconv::deconv_bulk
deconv_subgroups = compute.deconvolution.analysis(deconvolution = deconv_bulk,
corr = 0.7,
seed = 123,
file_name = "Tutorial",
return = TRUE) Subgroups composition can be extracted with:
deconv_subgroups[[3]]$B.cells
#> $B.cells_Subgroup.2.Iteration.1
#> [1] "DeconRNASeq_CBSX.HNSCC.scRNAseq_B.cells"
#> [2] "CBSX_CBSX.HNSCC.scRNAseq_B.cells"
#>
#> $B.cells_Subgroup.3.Iteration.1
#> [1] "Epidish_CBSX.Melanoma.scRNAseq_B.cells"
#> [2] "CBSX_CBSX.Melanoma.scRNAseq_B.cells"
#> [3] "DWLS_CBSX.Melanoma.scRNAseq_B.cells"
#>
#> $B.cells_Subgroup.4.Iteration.1
#> [1] "DeconRNASeq_CBSX.NSCLC.PBMCs.scRNAseq_B.cells"
#> [2] "CBSX_BPRNACan_B.cells"
#>
#> $B.cells_Subgroup.5.Iteration.1
#> [1] "Epidish_CBSX.NSCLC.PBMCs.scRNAseq_B.cells"
#> [2] "DWLS_CBSX.NSCLC.PBMCs.scRNAseq_B.cells"
#>
#> $B.cells_Subgroup.1.Iteration.2
#> [1] "DeconRNASeq_BPRNACan_B.cells" "DeconRNASeq_BPRNACanProMet_B.cells"
#>
#> $B.cells_Subgroup.2.Iteration.2
#> [1] "DeconRNASeq_CBSX.Melanoma.scRNAseq_B.cells"
#> [2] "B.cells_Subgroup.2.Iteration.1"
#>
#> $B.cells_Subgroup.3.Iteration.2
#> [1] "DeconRNASeq_CCLE.TIL10_B.cells" "DeconRNASeq_TIL10_B.cells"
#>
#> $B.cells_Subgroup.4.Iteration.2
#> [1] "CBSX_BPRNACan3DProMet_B.cells" "B.cells_Subgroup.4.Iteration.1"
#>
#> $B.cells_Subgroup.5.Iteration.2
#> [1] "CBSX_CBSX.NSCLC.PBMCs.scRNAseq_B.cells"
#> [2] "B.cells_Subgroup.5.Iteration.1"
#>
#> $B.cells_Subgroup.1.Iteration.3
#> [1] "B.cells_Subgroup.2.Iteration.2" "B.cells_Subgroup.4.Iteration.2"
#>
#> $B.cells_Subgroup.1.Iteration.4
#> [1] "B.cells_Subgroup.3.Iteration.1" "B.cells_Subgroup.1.Iteration.3"
deconv_subgroups[[3]]$Macrophages.M2
#> $Macrophages.M2_Subgroup.1.Iteration.1
#> [1] "Epidish_CCLE.TIL10_Macrophages.M2" "DWLS_CCLE.TIL10_Macrophages.M2"
#> [3] "CBSX_CCLE.TIL10_Macrophages.M2"
#>
#> $Macrophages.M2_Subgroup.2.Iteration.1
#> [1] "Epidish_TIL10_Macrophages.M2" "DWLS_TIL10_Macrophages.M2"
#>
#> $Macrophages.M2_Subgroup.4.Iteration.1
#> [1] "Epidish_LM22_Macrophages.M2" "DWLS_LM22_Macrophages.M2"
#> [3] "CBSX_LM22_Macrophages.M2"
#>
#> $Macrophages.M2_Subgroup.1.Iteration.2
#> [1] "DWLS_BPRNACan_Macrophages.M2"
#> [2] "DWLS_BPRNACan3DProMet_Macrophages.M2"
#> [3] "DWLS_BPRNACanProMet_Macrophages.M2"
deconv_subgroups[[3]]$Dendritic.cells
#> $Dendritic.cells_Subgroup.1.Iteration.1
#> [1] "Epidish_CBSX.HNSCC.scRNAseq_Dendritic.cells"
#> [2] "DWLS_CBSX.HNSCC.scRNAseq_Dendritic.cells"
#> [3] "CBSX_CBSX.HNSCC.scRNAseq_Dendritic.cells"Reduced deconvolution matrix:
head(subgroups[[1]][,sample(colnames(subgroups[[1]]), 10)])
#> Quantiseq_T.cells.non.regulatory
#> SAM7f0d9cc7f001 0.01070804
#> SAM4305ab968b90 0.00000000
#> SAMcf018fee2acd 0.00000000
#> SAMcc4675f394a1 0.00000000
#> SAM49f9b2e57aa5 0.00000000
#> SAM2e7aa8fa0ab3 0.00000000
#> Epidish_CBSX.NSCLC.PBMCs.scRNAseq_NKT.cells
#> SAM7f0d9cc7f001 0.06177058
#> SAM4305ab968b90 0.20596451
#> SAMcf018fee2acd 0.10081728
#> SAMcc4675f394a1 0.16370762
#> SAM49f9b2e57aa5 0.18693083
#> SAM2e7aa8fa0ab3 0.05430822
#> DWLS_BPRNACanProMet_Macrophages.M1
#> SAM7f0d9cc7f001 0.000000000
#> SAM4305ab968b90 0.000000000
#> SAMcf018fee2acd 0.000000000
#> SAMcc4675f394a1 0.003182723
#> SAM49f9b2e57aa5 0.000000000
#> SAM2e7aa8fa0ab3 0.000000000
#> Epidish_CBSX.NSCLC.PBMCs.scRNAseq_CD8.cells
#> SAM7f0d9cc7f001 0
#> SAM4305ab968b90 0
#> SAMcf018fee2acd 0
#> SAMcc4675f394a1 0
#> SAM49f9b2e57aa5 0
#> SAM2e7aa8fa0ab3 0
#> DWLS_CBSX.NSCLC.PBMCs.scRNAseq_NK.cells
#> SAM7f0d9cc7f001 0.000000
#> SAM4305ab968b90 0.000000
#> SAMcf018fee2acd 0.000000
#> SAMcc4675f394a1 0.000000
#> SAM49f9b2e57aa5 0.000000
#> SAM2e7aa8fa0ab3 0.118806
#> DeconRNASeq_CBSX.NSCLC.PBMCs.scRNAseq_NKT.cells
#> SAM7f0d9cc7f001 0.1332183
#> SAM4305ab968b90 0.1410772
#> SAMcf018fee2acd 0.1134181
#> SAMcc4675f394a1 0.1362632
#> SAM49f9b2e57aa5 0.1496804
#> SAM2e7aa8fa0ab3 0.1424695
#> CBSX_CBSX.NSCLC.PBMCs.scRNAseq_NK.cells
#> SAM7f0d9cc7f001 0.00000000
#> SAM4305ab968b90 0.00000000
#> SAMcf018fee2acd 0.00000000
#> SAMcc4675f394a1 0.00000000
#> SAM49f9b2e57aa5 0.00000000
#> SAM2e7aa8fa0ab3 0.02834543
#> Endothelial_Subgroup.1.Iteration.1
#> SAM7f0d9cc7f001 0.05480971
#> SAM4305ab968b90 0.02591334
#> SAMcf018fee2acd 0.05501908
#> SAMcc4675f394a1 0.01745065
#> SAM49f9b2e57aa5 0.00000000
#> SAM2e7aa8fa0ab3 0.00000000
#> DeconRNASeq_CBSX.Melanoma.scRNAseq_CAF
#> SAM7f0d9cc7f001 0.4362575
#> SAM4305ab968b90 0.2159553
#> SAMcf018fee2acd 0.2789918
#> SAMcc4675f394a1 0.3163672
#> SAM49f9b2e57aa5 0.2855141
#> SAM2e7aa8fa0ab3 0.4346621
#> DeconRNASeq_LM22_Macrophages.M2
#> SAM7f0d9cc7f001 0.025825608
#> SAM4305ab968b90 0.000000000
#> SAMcf018fee2acd 0.027464529
#> SAMcc4675f394a1 0.081372076
#> SAM49f9b2e57aa5 0.000000000
#> SAM2e7aa8fa0ab3 0.004871071If your deconvolution matrix contains non-standard cell types (see
README), specify them using cells_extra to ensure proper
subgrouping. If not, they are going to be discarded automatically.
deconv_subgroups = compute.deconvolution.analysis(deconvolution = deconv_pseudo,
corr = 0.7,
seed = 123,
return = TRUE,
cells_extra = c("Mural.cells", "Myeloid.cells"),
file_name = "Tutorial") Deconvolution dictionary
The deconvolution dictionary step integrates deconvolution features with pathway activity information to provide a functional interpretation of each cell-type–specific component. This process identifies groups of pathways that show coordinated behavior across samples (e.g., immunoactive vs. immunosuppressive signaling) and assigns each deconvolution feature to one of these pathway clusters based on correlation patterns. The resulting dictionary can then be used to understand whether a cell-type feature is more associated with activation or suppression programs.
For this, we start from normalized gene expression data, compute pathway activity scores (see CellTFusion package), and link them to deconvolution results obtained in previous steps. Finally, we use the deconvolution_dictionary() function to classify each deconvolution feature according to its dominant pathway cluster.
counts = multideconv::raw_counts
counts.norm = ADImpute::NormalizeTPM(counts, log = TRUE)
pathways = CellTFusion::compute.pathway.activity(counts.norm)
deconv = multideconv::deconv_bulk[rownames(multideconv::deconv_bulk)%in%colnames(counts),]
deconv_subgroups = compute.deconvolution.analysis(deconvolution = deconv,
corr = 0.7,
seed = 123,
file_name = "Tutorial",
return = TRUE)
deconv_subgroups = deconvolution_dictionary(deconv_subgroups, pathways)
head(deconv_subgroups[[1]][,1:5])
print(deconv_subgroups[["States"]])Replicate deconvolution subgroups in an independent set
Cell subgroup identification through deconvolution is cohort-specific, as it relies on correlation patterns across samples. This means that subgroup definitions may vary across different splits or datasets. If you aim to replicate the same subgroups identified in one dataset onto another (e.g., for model validation), you can use the following function.
The function below reconstructs and applies the subgroup signatures derived from a previous deconvolution, making it especially useful when transferring learned patterns across datasets—such as when training and evaluating machine learning models.
deconv_1 = deconv_bulk[1:100,]
deconv_2 = deconv_bulk[101:192,]
deconv_subgroups = compute.deconvolution.analysis(deconvolution = deconv_1,
corr = 0.7,
seed = 123,
file_name = "Tutorial",
return = FALSE)
deconv_subgroups_replicate = replicate_deconvolution_subgroups(deconv_subgroups,
deconv_2)Machine learning models using deconvolution subgroups
The deconvolution subgroups generated by multideconv can
be used as input features for training machine learning (ML) models.
However, since these subgroups are derived based on sample-level
correlations, special care is needed to avoid data leakage. If you
compute the full subgroup matrix on the entire dataset before splitting
into training and test sets (or before performing k-fold
cross-validation), the model may indirectly access information from the
test set during training.
To address this issue, we provide the function
prepare_multideconv_folds(), which ensures proper
separation between training and test data during the computation of
deconvolution subgroups. This function constructs ML folds in a way that
subgroup features are learned only from the training data of each fold
and then projected onto the corresponding test fold out.
Below is an example usage of the multideconv subgroups
using pipeML, an R package that includes direct flexibility
into k-fold construction. We will add our custom function to compute the
folds of the model using multideconv in the argument
fold_construction_fun. For more information about the
functions arguments visit the documentation of pipeML :
library(pipeML)
# traitData_train: Subset of clinical data used for training
# traitData_test: Subset of clinical data used for testing
# deconv_train: Deconvolution of samples used for training
# deconv_test: Deconvolution of samples used for testing
# Training in deconv subgroups
res = pipeML::compute_features.training.ML(deconv_train,
traitData_train$Response,
trait.positive = "R",
metric = "AUROC",
stack = F,
k_folds = 5,
n_rep = 10,
feature.selection = F,
LODO = F,
ncores = 3,
return = F,
fold_construction_fun = prepare_multideconv_folds)
# Replicate deconvolution subgroups
dt_test = replicate_deconvolution_subgroups(res$Custom_output$Processed_deconvolution,
deconv_test)
# Predict in deconv subgroups
pred = pipeML::compute_prediction(res,
dt_test,
traitData_test$Response,
trait.positive = "R",
stack = F,
maximize = "Accuracy",
return = T)NOTE: multideconv is built on top of
existing frameworks and makes extensive use of the R packages
immunedeconv (Sturm et al. (2019)) and
omnideconv (Dietrich et al. (2024)). If you use
multideconv in your work, please cite our package along
with these foundational packages. We also encourage you to cite the
individual deconvolution algorithms you employ in your analysis.
| method | license | citation |
|---|---|---|
| quanTIseq | free (BSD) | Finotello, F., Mayer, C., Plattner, C., Laschober, G., Rieder, D., Hackl, H., …, Sopper, S. (2019). Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome medicine, 11(1), 34. https://doi.org/10.1186/s13073-019-0638-6 |
| EpiDISH | free (GPL 2.0) | Zheng SC, Breeze CE, Beck S, Teschendorff AE (2018). “Identification of differentially methylated cell-types in Epigenome-Wide Association Studies.” Nature Methods, 15(12), 1059. https://doi.org/10.1038/s41592-018-0213-x |
| DeconRNASeq | free (GPL-2) | joseph.szustakowski@novartis.com TGJDS (2025). DeconRNASeq: Deconvolution of Heterogeneous Tissue Samples for mRNA-Seq data. doi:10.18129/B9.bioc.DeconRNASeq, R package version 1.50.0, https://bioconductor.org/packages/DeconRNASeq |
| AutoGeneS | free (MIT) | Aliee, H., & Theis, F. (2021). AutoGeneS: Automatic gene selection using multi-objective optimization for RNA-seq deconvolution. https://doi.org/10.1101/2020.02.21.940650 |
| BayesPrism | free (GPL 3.0) | Chu, T., Wang, Z., Pe’er, D. et al. Cell type and gene expression deconvolution with BayesPrism enables Bayesian integrative analysis across bulk and single-cell RNA sequencing in oncology. Nat Cancer 3, 505–517 (2022). https://doi.org/10.1038/s43018-022-00356-3 |
| Bisque | free (GPL 3.0) | Jew, B., Alvarez, M., Rahmani, E., Miao, Z., Ko, A., Garske, K. M., Sul, J. H., Pietiläinen, K. H., Pajukanta, P., & Halperin, E. (2020). Publisher Correction: Accurate estimation of cell composition in bulk expression through robust integration of single-cell information. Nature Communications, 11(1), 2891. https://doi.org/10.1038/s41467-020-16607-9 |
| BSeq-sc | free (GPL 2.0) | Baron, M., Veres, A., Wolock, S. L., Faust, A. L., Gaujoux, R., Vetere, A., Ryu, J. H., Wagner, B. K., Shen-Orr, S. S., Klein, A. M., Melton, D. A., & Yanai, I. (2016). A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. In Cell Systems (Vol. 3, Issue 4, pp. 346–360.e4). https://doi.org/10.1016/j.cels.2016.08.011 |
| CIBERSORTx | free for non-commerical use only | Newman, A. M., Liu, C. L., Green, M. R., Gentles, A. J., Feng, W., Xu, Y., Hoang, C. D., Diehn, M., & Alizadeh, A. A. (2015). Robust enumeration of cell subsets from tissue expression profiles. Nature Methods, 12(5), 453–457. https://doi.org/10.1038/s41587-019-0114-2 |
| CPM | free (GPL 2.0) | Frishberg, A., Peshes-Yaloz, N., Cohn, O., Rosentul, D., Steuerman, Y., Valadarsky, L., Yankovitz, G., Mandelboim, M., Iraqi, F. A., Amit, I., Mayo, L., Bacharach, E., & Gat-Viks, I. (2019). Cell composition analysis of bulk genomics using single-cell data. Nature Methods, 16(4), 327–332. https://doi.org/10.1038/s41592-019-0355-5 |
| DWLS | free (GPL) | Tsoucas, D., Dong, R., Chen, H., Zhu, Q., Guo, G., & Yuan, G.-C. (2019). Accurate estimation of cell-type composition from gene expression data. Nature Communications, 10(1), 2975. https://doi.org/10.1038/s41467-019-10802-z |
| MOMF | free (GPL 3.0) | Xifang Sun, Shiquan Sun, and Sheng Yang. An efficient and flexible method for deconvoluting bulk RNAseq data with single-cell RNAseq data, 2019, DOI: 10.5281/zenodo.3373980 |
| MuSiC | free (GPL 3.0) | Wang, X., Park, J., Susztak, K., Zhang, N. R., & Li, M. (2019). Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nature Communications, 10(1), 380. https://doi.org/10.1038/s41467-018-08023-x |
| SCDC | (MIT) | Dong, M., Thennavan, A., Urrutia, E., Li, Y., Perou, C. M., Zou, F., & Jiang, Y. (2020). SCDC: bulk gene expression deconvolution by multiple single-cell RNA sequencing references. Briefings in Bioinformatics. https://doi.org/10.1093/bib/bbz166 |